This post was originally posted on June 3rd 2020.
In case the name doesn’t give it away, multiplex real-time PCR involves amplifying multiple DNA or RNA targets simultaneously in a single PCR reaction. This requires the presence of a specific pair of primers and a complementary DNA-binding probe for each target under study.
Multiplex PCR arose in the late 1980s as a rapid method to detect mutations in the gene responsible for Duchenne muscular dystrophy. Today, the technique is widely used as a research tool for gene expression analysis and genomics, and it also has clinical applications in diagnostics, viral load monitoring, genetic testing and more.
What Are the Benefits of Multiplex Real-Time PCR?
Setting up a multiplex PCR experiment requires greater effort than a singleplex real-time PCR experiment. However, once the setup is in place, the researcher can enjoy many benefits including:
- Long-term time and cost savings – optimised setups may yield savings on personnel costs, as well as on dNTPS and other reagents
- Increased throughput compared to regular real-time PCR
- Reduced impact of pipetting errors because there are fewer pipetting steps
- Superior data normalisation because of the possibility to add an internal reference to each PCR reaction
In some fields, the biggest attraction of all is the possibility to get more data out of very limited amounts of staring material, e.g., raw material is scare or nucleic acid yields are low. For research projects that routinely focus on the same group of genes, multiplexing is also an attractive option because in addition to the benefits outlined above it also permits higher throughput than is possible with singleplex PCR reactions.
Multiplex Real-Time PCR Workflow
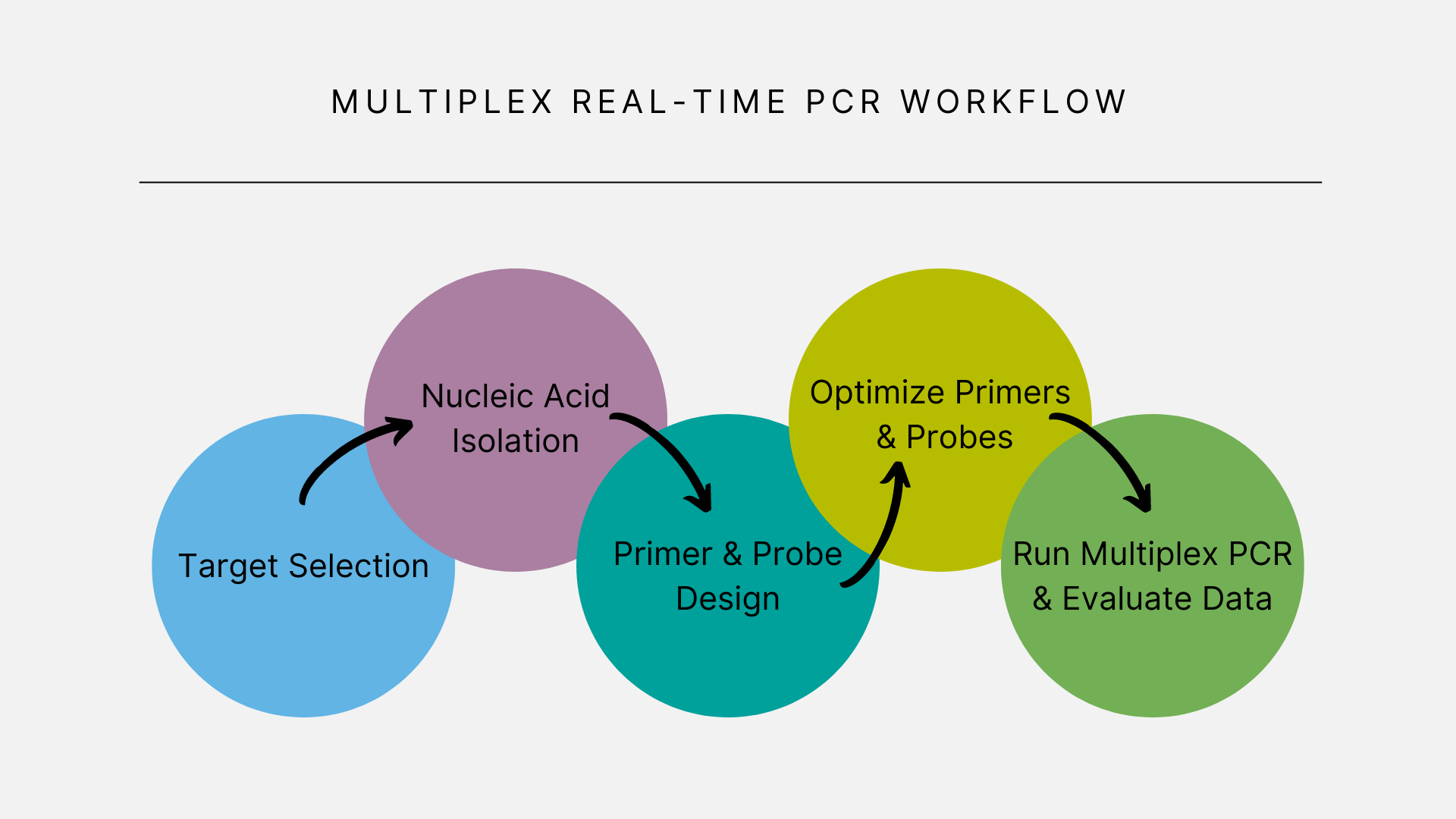
At a glance, the typical multiplex workflow doesn’t appear very different from a singleplex real-time PCR workflow. However, there are some key differences. Let’s take a look.
Choosing a Reporter
In Part 1 of our real-time PCR series, we explained how real-time PCR differs from conventional or end-point PCR. To recap, end-point PCR reactions are typically analysed qualitatively by agarose gel electrophoresis. While it is sometimes possible to make estimates about amplicon concentration by comparing DNA bands to a marker of known concentration, this approach is not suitable for applications that require precise quantification of DNA or RNA targets.
In real-time PCR, quantitative monitoring of template amplification is made possible by the presence of a fluorescent reporter molecule in each PCR reaction that produces a fluorescent signal as the amount of PCR amplicon increases.
There are two main choices when it comes to fluorescent reporters for real-time PCR:
- Non-specific DNA-binding dyes that intercalate dsDNA in a sequence-independent manner, e.g., SYBR green
- Sequence-specific fluorescence quencher probes, e.g., TaqMan. These are custom-designed with complementarity to the target sequence and generally consist of 3 components: a DNA-binding region, a 5’-fluorophore and a 3’-quencher. During PCR extension, newly formed target DNA is bound by the probe. As Taq polymerase extends along the template, it eventually meets the 3’-quencher and cleaves it via its inherent 5’-3’ exonuclease activity. With the quencher on the loose, the fluorophore emits a measurable signal.
Probing for Success!
Given the non-specific nature of SYBR Green and other dyes in this category, they should be used with extreme caution in multiplex PCR setups. Theoretically, it should be possible to discriminate between multiple targets if the melting temperatures (Tm) of their amplicons are different enough, but in general this is seen as a risky approach especially when a more reliable option exists, namely the fluorescent quencher probes.
A number of online tools exist for probe and primer design with options for multiplex PCR. In addition, probe manufacturers and suppliers are usually available to provide solid advice about probe design and experimental setup. For now, here are a few general tips to start you off:
- Aim to design probes with melting temperatures approx. 7-8 °C higher than the Tm of the corresponding primer pair. This allows the probe to bind the target first, minimising the risk of target sequences escaping detection.
- Bear in mind that competition between targets for reaction components may result in detection of highly abundant targets while low abundance targets are misrepresented. The only way to mitigate this risk is to carefully design primer pairs and optimise reaction conditions (annealing temperature, cycle number, etc.) so that each PCR reaction in the multiplex setup operates at equal efficiency. Choosing a PCR mastermix that is optimised for multiplex PCR may also help reduce the competition for reagents.
- The number of fluorescent labels available and overlap in their spectral profiles may make experimental planning a challenge. Choose fluorophores that are spectrally distinct, and if possible, ensure that your most abundant target is detected by the least intense dye and vice-versa.
- A standard real-time PCR instrument has 4 or 5 channels available for colour detection. If you wish to assess more than 4 or 5 targets at a time (check what your instrument can do), you may want to check out the research literature for alternative setups based on bead- or chemically-labelled probes for the possibility to include larger target numbers.
- Validate your multiplex setup by running singleplex and multiplex reactions side by side on the same PCR plate before proceeding with your multiplex experiments ‘for real’.
What About Pitfalls?
There are a few major pitfalls associated with multiplexing that are worth highlighting again. Firstly, competition for reagents and undesired interactions between components within the multiplexed PCR reaction can greatly impact result accuracy. These include unwanted interactions between primers and other primers, primers and probes as well as PCR inhibition. The more targets under investigation in a single reaction the more difficult it becomes to mitigate these issues, so careful primer and probe design and extensive validation are a must.
A particularly risky scenario can arise when one of the targets under investigation is very abundant in relation to the others. This target will likely reach its plateau phase of amplification much earlier that a low abundance target, perhaps even before the low abundance target has undergone detectable amplification. The risk is that the reagents will then become limiting for the low abundance target, resulting in a Ct value that misrepresents its abundance, leaving you with dodgy results. The solution for this is primer limitation. In simple terms, this involves limiting the primer concentration for the most abundant target, so that its reaches it plateau more quickly (because primers become depleted). This should leave ample reagents for the remaining targets to be amplified and represented accurately.
Planning and Validation Are Key!
Beyond the tips we’ve shared here, standard considerations for real-time PCR such as housekeeping gene selection, internal standard selection, primer design and optimisation, reaction setup (e.g., plate layout), experimental controls and replicates, and post-run quality control are of equal importance in multiplex PCR. All of these considerations are covered in detail in our real-time PCR series.
For questions relating to multiplex PCR planning, setup and validation, we encourage you to get in touch with us so that we can advise you based on your exact circumstances. You can find contact details for your local representative here.